Mapping brain circuits – deciphering the connectome
Neuroscientists want to understand how tangles of neurons produce complex behaviours, but even the simplest networks defy understanding.
An exciting and profoundly consequential endeavor of neuroscience that may be realized this century is the complete mapping of the human brain – the connectome. However, just like the mapping of the human genome, the usefulness of such data is limited if there is no contextual understanding of what it means. The mapping of the human genome revealed some surprising results, but it was met with limited success in previously purported claims such as enabling greater prediction of genetic predispositions for particular diseases. In part because even with the genomic data it is not yet understood exactly how gene sequences correlate to certain traits — they are defined by complex molecular circuitry that do not have straight-forward and easily decipherable “rules of expression”, i.e. there is little to no predictive power.
Similarly, as discussed in a recent Nature news report, “the resulting neural-network diagrams emerging from mapping brain circuits [in a variety of species] are yielding surprises — showing, for example, that a brain can use one network in multiple ways to create the same behaviors. But understanding even the simplest of circuits presents a host of challenges. Circuits vary in layout and function from animal to animal. The systems have redundancy that makes it difficult to pin one function to one circuit. Plus, wiring alone doesn't fully explain how circuits generate behaviors; other factors, such as neurochemicals, have to be considered.”
As Florian Engert, who is putting together an atlas of the zebrafish brain at Harvard University in Cambridge, Massachusetts, puts it “What do you even mean when you say you understand how something works? If you map it out, you haven't really understood anything.”
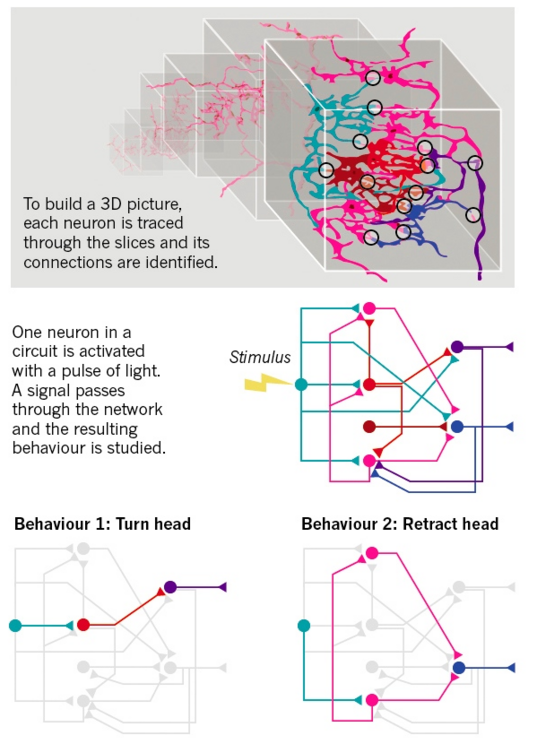
William Brown, a biophysicist with the Resonance Science Foundation, has stated that the complications in ascertaining a clear and understandable picture from all the (highly important) data coming from mapping genomes, transcriptomes, proteomes, and connectomes may be due to the bottom-up reductionist approach that attempts to understand biological systems as the sum of action of their parts. Yet, such systems are more than the sum of their parts—they are necessarily highly integrated and synergetic, which may require a complimentary top-down approach that evaluates the ”big picture”—in this case the unified view of the organism that has emergent properties that can only be understood in the context of the synergetic action of the system as a whole. Such approaches have seen remarkable success with researchers such as Dr. Miguel Nicolelis, who has decoded the spatiotemporal global activity of electromagnetic patterns of the brain to describe function and even enable mobility of paraplegics with neuroprosthetic devices.

